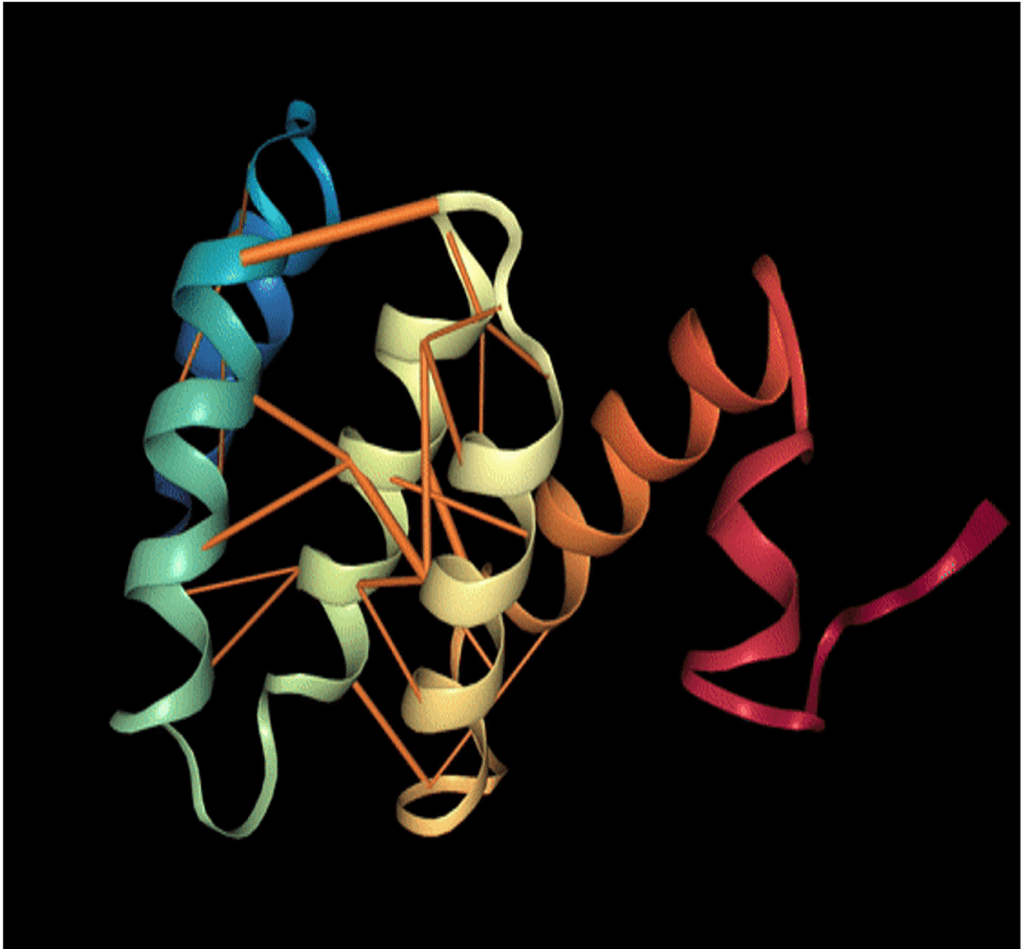For more than a year now, the world has been dealing with a Sars-Cov-2 virus. Its speed of spread and the diversity of geographical areas affected quickly classified it as a pandemic. To deal with this health crisis, research has multiplied in all scientific sectors with a double challenge: to find a treatment but also to limit the spread of the epidemic. This is where Natural Language Processing came into play in the fight against COVID-19. What role could the NLP have played in the development of a treatment ? How did it limit the negative consequences of the pandemic ? How was it used to estimate the impact of the crisis? We explain everything in this article.
By Guillaume SCHALLER, Data Scientist at LittleBigCode

The funds allocated to research combined with public and private collaboration made it possible to combat a pandemic of this scale in an unprecedented way. As an example of this global effort, on 1 February 2021, there were over 105,000 scientific publications according to the COVID-19 Primer website. At the same time, artificial intelligence has achieved very convincing results for its use in our daily lives, facilitated in this sense by the amount of accessible data and the computing capacity of computers.
Natural Language Processing (NLP), a major branch of artificial intelligence, has greatly contributed to this revolution through its wide range of applications (automatic translation, summarisation, sentiment extraction). NLP models have also improved greatly in recent years. For example, the latter are often assessed on GLUE (General Language Understanding Evaluation), which is a collection of various linguistic problems (inference, paraphrasing, similarity between texts, etc.): Microsoft’s DeBERTa obtained an overall score of 90.8, whereas the baseline was only 70 two years ago. This is why artificial intelligence has made it possible to respond to this health crisis in an unprecedented way.
1/ NLP to decipher the language of life
The first use case for the NLP was in the search for a treatment for COVID-19. How ? Using the same models that are used to analyse our language, the scientists studied the protein sequences in order to determine the genetic skeleton of the virus within a few weeks.
In simple terms, a protein is a variable length sequence of about 20 amino acids. From this point of view, proteins can be considered as any language with its alphabet (the amino acids) to form sentences (the primary sequence of the protein). While it is almost impossible for us humans to learn to speak the protein, NLP models, by analysing billions of protein sequences associated with their property, can understand its grammar.
Public databases such as Uniprot have made these studies possible. However, these data are extremely costly to label as they require a great deal of knowledge. The models used to make use of this wealth of data are therefore self-supervised learning models.
2/ Models based on Transformers
What is it about? The idea of the Transformers is the same as that which led to the birth of the RNN (Recurrent Neural Network) models. The processing of the input should depend on the previous input. The main limitation of these RNN models is that they have a very limited context window, which prevents them from understanding the relationships that may exist between entries that are far apart in the sequence. Given that proteins have average sequences that exceed 300 tokens, it’s normal that RNNs are little used to study protein sequences.
A) Transformer model
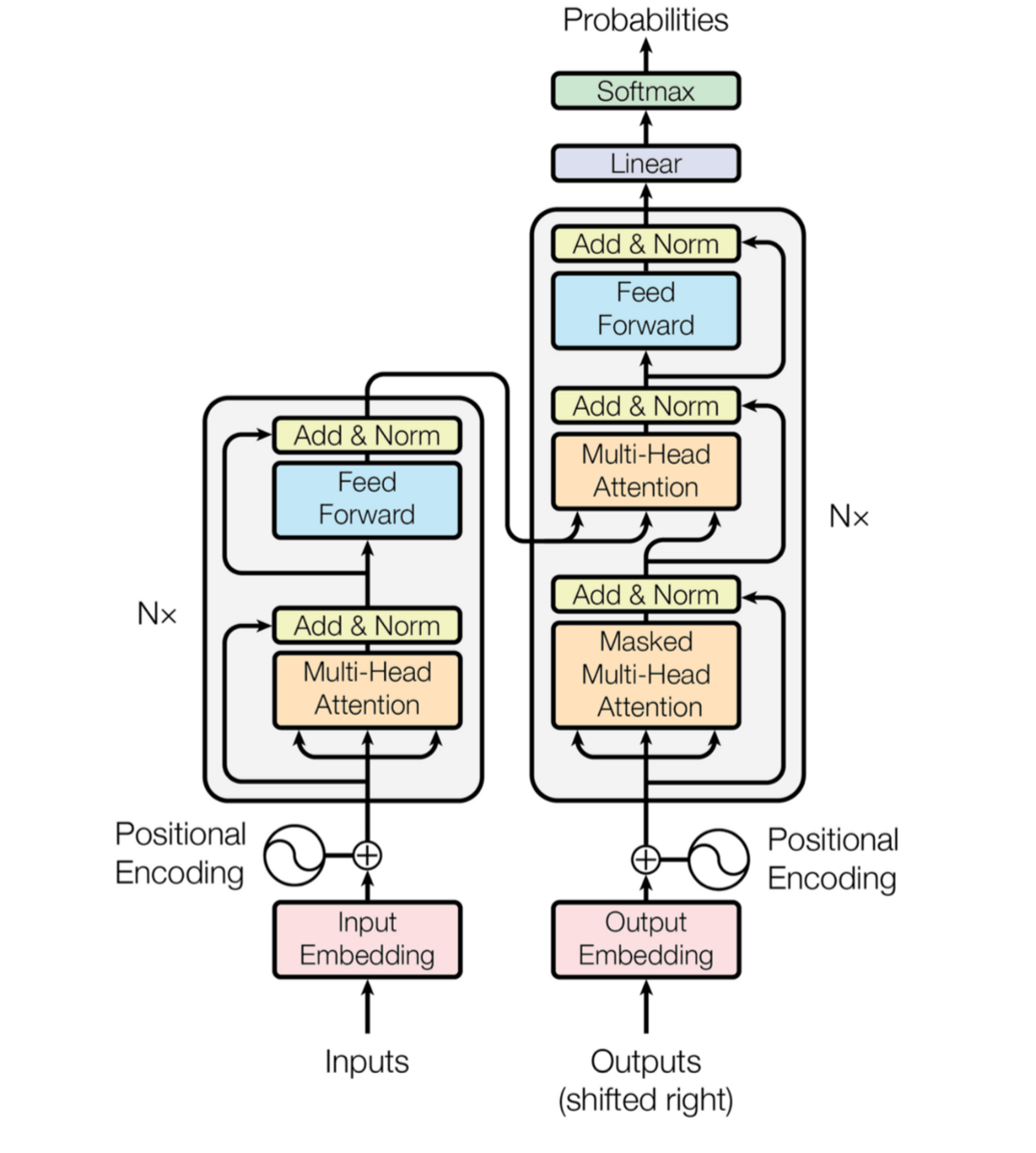
Transformers models of the scientific article Attention is all you need study inputs according to a context but without any layer of recurrent neurons. These models use layers of attention to understand the dependencies between inputs at different positions.
FIGURE 1 : Diagram of a transformer (source : Attention is all you need)
As you can see from the diagram, the encoder and decoder stack layers of multi-head attention and feed-forward associated with word position encoding. Objective: to help the model maintain an understanding of the sequence. In addition, the use of normalisation layers can be noted in order to avoid the “vanishing gradient” due to the large number of matrix products in the back-propagation.
B) The attention mechanism in protein sequences
Proteins, as explained earlier, are sequences of amino acids that form a primary sequence. The interactions between the different blocks of a sequence create shapes (helices…) which are the secondary structure of a protein. Finally, these forms interact to form the tertiary structure of the protein.
In order to obtain more interpretation in the results obtained by NLP models on proteins, Rajesh Bhasin and Abhishek Kumar, in their article ProVis : An Anaglyph based Visualization Tool for Protein Molecules, coupled the model attention weight of the individual amino acids with the tertiary structure of the protein. Indeed, each attention layer produces a probability distribution w where represents the attention weight between the token and . The greater the weight, the more dependent these tokens are.
In this scientific paper, it’s realised that amino acids that are far apart in the primary sequence input to the model can have a high dependency. This is explained by their proximity in 3D after the formation of the tertiary structure of the protein. Thus, without having any clue about the physical structure of the protein, the NLP models were able to capture the strong interaction between two spatially close amino acids.
FIGURE 2: 3D protein with orange attention mechanisms from a model trained on their primary 1D sequences. The orange cylinders are the DD attention weights whose diameter is proportional to the value.
C) Evaluation of models
But visualizing attention mechanisms is not everything. We also need to be able to quickly evaluate protein language modelling with metrics that make sense, both at the NLP level and also at the biological level.
Initially, as with an ordinary language, the models can be evaluated on their perplexity. This score, which is a function of the cross-entropy of a sequence, measures the generalisability of the grammar of a language. Indeed, if the perplexity of a word is high, it means that the model finds it completely unsuitable in the sequence. Whereas a model with low perplexity will have learned the nuances of a language. So seeing a new word with a similar meaning will not surprise him.
This is why we are looking for language representation models with low perplexity. Another possible metric is based on the accuracy of a model in predicting a token that has been masked.
Regarding metrics of interest to biologists, a language representation model can be evaluated on a corpus of bioinformatics tasks. Presented in the scientific article Evaluating Protein Transfer Learning with TAPE | bioRxiv, this corpus is an analogy with the method of evaluating NLP models on a classical GLUE language.
TAPE provides 5 tasks with their associated datasets to benchmark a model against the state of the art. These 5 tasks (Secondary Structure Prediction, Contact Prediction, Remote Homology Detection, Fluorescence Landscape Prediction and Stability Landscape Prediction) all address protein modelling issues.
For example, the Secondary Structure Prediction task aims to assess the ability of a model that has only studied primary sequences to understand properties related to the secondary form of proteins. This task is therefore a sequence-to-sequence problem in which the model must predict for each amino acid its form in the second structure (three possible labels: helix, strand or other).
The Stability Landscape Prediction task is also a sequence-to-sequence regression problem that aims to test whether models learn to measure protein stability. This is extremely useful for research into the creation of functional proteins, as one can easily keep the mutations that are most likely to remain stable.
D) Protein generation for treatment research
The interest of protein generation is above all represented by the time saving it generates. Previously, the creation of new protein sequences was based on making mutations in parts of the protein sequence. The idea was to obtain a desired property with biological techniques such as direct evolution.
However, for a sequence of length L consisting of m amino acids, there are O( ) possibilities of mutations. Each change is time consuming and expensive.
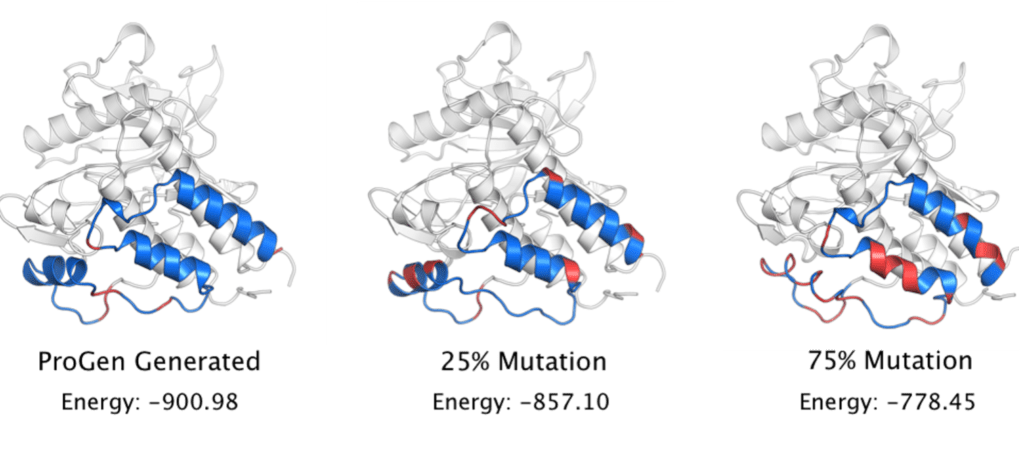
Articles such as ProGen: Using AI to Generate Proteins (einstein.ai) propose to imitate this. They model the language of proteins and then select protein sequences known to have certain desired properties. Then they hide parts of the sequence and generate the missing sequences with their model. Generated candidates with the best biophysical properties can then be recreated in the laboratory. They demonstrated that their generated sequences were functional because they retained the same secondary structures as the original sequences and minimised internal energy. To test their results against random mutations, they then created baselines by randomly modifying 25 and 75% of the sequence with random amino acids.
FIGURE 3 : Sequence of generated proteins. The conservation of secondary structure is highlighted by the internal energy (low in BLUE, high in RED). Source : Blog Einstein
In this first part of the article, we saw how NLP could be used to identify the mechanism of the virus and help develop a treatment. In a second part, we will see how natural language processing has also helped to limit the consequences of the pandemic.